Mapping Quantitative Trait Loci (QTL): Unraveling the Genetic Structure of Complicated Traits
Associated Articles: Mapping Quantitative Trait Loci (QTL): Unraveling the Genetic Structure of Complicated Traits
Introduction
On this auspicious event, we’re delighted to delve into the intriguing matter associated to Mapping Quantitative Trait Loci (QTL): Unraveling the Genetic Structure of Complicated Traits. Let’s weave attention-grabbing info and supply contemporary views to the readers.
Desk of Content material
Mapping Quantitative Trait Loci (QTL): Unraveling the Genetic Structure of Complicated Traits

Quantitative traits, not like discrete traits like flower colour (pink or white), exhibit steady variation inside a inhabitants. Peak, weight, yield, illness resistance – these are all examples of quantitative traits influenced by the mixed results of a number of genes (polygenes) and environmental components. Understanding the genetic foundation of those traits is essential in numerous fields, from agriculture to human well being. Mapping quantitative trait loci (QTL) is a robust statistical technique employed to establish genomic areas related to the variation in quantitative traits. This text delves into the ideas, methodologies, and purposes of QTL mapping.
Understanding Quantitative Traits and Their Genetic Structure:
The phenotypic expression of a quantitative trait is the results of a fancy interaction between genotype and surroundings. The genotype refers back to the particular person’s genetic make-up at a number of loci, every contributing a small impact to the general phenotype. These genes, referred to as quantitative trait genes (QTGs), usually work together with one another (epistasis) and with environmental components (GxE interplay). The environmental contribution can embody components akin to temperature, nutrient availability, or illness strain. The problem of QTL mapping lies in disentangling the contributions of those a number of interacting components to the noticed phenotypic variation.
The Rules of QTL Mapping:
QTL mapping depends on the precept of linkage disequilibrium (LD). LD refers back to the non-random affiliation between alleles at completely different loci on a chromosome. If a QTG is carefully linked to a marker locus (a recognized DNA sequence variation), the alleles on the marker and the QTG will are usually inherited collectively extra usually than anticipated by probability. By analyzing the segregation of marker alleles and the phenotypic values in a inhabitants, we will infer the presence and site of QTGs.
Approaches to QTL Mapping:
A number of approaches exist for QTL mapping, every with its strengths and limitations. Two main strategies are generally used:
1. Interval Mapping: This technique makes use of a single marker at a time to evaluate its affiliation with the trait. It fashions the probability of a QTL being positioned between two adjoining markers primarily based on the noticed marker genotypes and phenotypes. Whereas computationally much less intensive than different strategies, interval mapping can undergo from diminished energy when QTLs are carefully spaced.
2. Composite Interval Mapping (CIM): CIM addresses the constraints of interval mapping by incorporating extra markers as covariates within the statistical mannequin. These extra markers management for the results of different QTLs, thereby rising the precision and energy of QTL detection. This reduces the probability of false positives brought on by linkage between markers and different QTLs.
3. A number of Interval Mapping (MIM): MIM concurrently maps a number of QTLs, explicitly contemplating their epistatic interactions. This technique is especially helpful for dissecting complicated traits influenced by quite a few interacting genes. Nevertheless, the computational calls for of MIM are considerably greater in comparison with interval and composite interval mapping.
4. Genome-Huge Affiliation Research (GWAS): Whereas not strictly a QTL mapping strategy, GWAS makes use of an identical precept to establish single nucleotide polymorphisms (SNPs) related to quantitative traits. GWAS sometimes makes use of bigger populations and a denser marker protection in comparison with conventional QTL mapping research, permitting for greater decision mapping. Nevertheless, GWAS depends on the belief of LD between SNPs and QTGs, which may be restricted by the extent of LD within the inhabitants.
Experimental Designs and Knowledge Necessities:
The success of QTL mapping depends closely on the experimental design and the standard of the information. Widespread experimental designs embody:
-
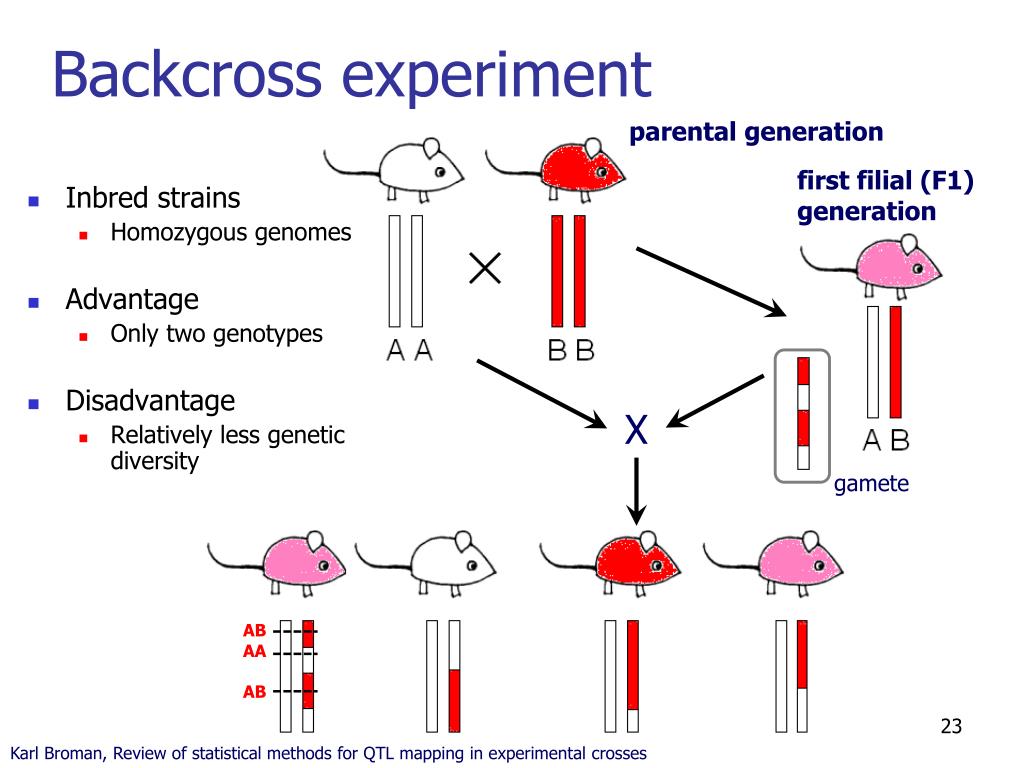
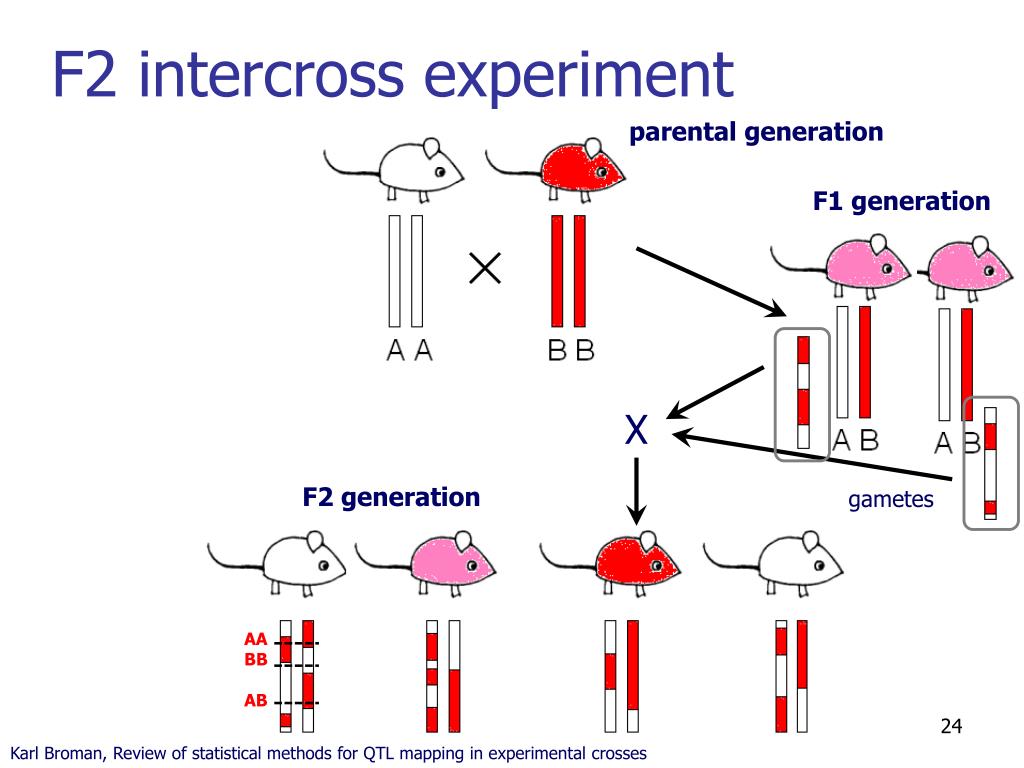
Biparental populations: These populations are derived from a cross between two homozygous dad and mom, akin to F2 populations (second filial era) or recombinant inbred strains (RILs). The segregation of alleles in these populations gives the idea for QTL detection.
-
Multiparental populations: These populations are derived from a number of parental strains, providing higher genetic variety and probably rising the facility to detect QTLs with small results.
-
Affiliation mapping populations: These populations make the most of present pure populations or various germplasm collections. They leverage present LD within the inhabitants to establish QTLs.
The information required for QTL mapping embody:
-
Genotypic knowledge: This entails marker genotypes for a set of people, sometimes SNPs or different molecular markers distributed throughout the genome.
-
Phenotypic knowledge: This consists of quantitative trait measurements for a similar people. Correct and dependable phenotypic knowledge are essential for correct QTL mapping.
Statistical Software program and Evaluation:
A number of statistical software program packages can be found for performing QTL mapping, together with:
-
QTL Cartographer: A broadly used package deal providing numerous mapping strategies, together with interval mapping and CIM.
-
R/qtl: An R package deal offering a complete set of features for QTL mapping and evaluation.
-
WinQTL Cartographer: A user-friendly Home windows-based program for QTL mapping.
The evaluation sometimes entails:
-
Linkage map building: Making a genetic map displaying the relative positions of markers on chromosomes.
-
QTL detection: Figuring out genomic areas displaying vital affiliation with the trait utilizing statistical exams.
-
QTL impact estimation: Estimating the magnitude and route of the impact of every QTL on the trait.
-
QTL interplay evaluation: Investigating the interactions between QTLs and environmental components.
Purposes of QTL Mapping:
QTL mapping has discovered large purposes in numerous fields:
-
Agriculture: Figuring out genes controlling yield, illness resistance, and different agronomically vital traits in crops and livestock. This info can be utilized in marker-assisted choice (MAS) to enhance breeding effectivity.
-
Human genetics: Mapping genes related to complicated human ailments, akin to diabetes, coronary heart illness, and most cancers. This helps to know the genetic structure of those ailments and develop higher diagnostic and therapeutic methods.
-
Evolutionary biology: Finding out the genetic foundation of adaptation and phenotypic evolution in pure populations. QTL mapping can reveal the genes answerable for adaptation to particular environments.
-
Forestry: Enhancing the expansion fee, wooden high quality, and illness resistance of bushes.
-
Animal breeding: Enhancing productiveness and illness resistance in livestock.
Challenges and Future Instructions:
Regardless of its energy, QTL mapping faces a number of challenges:
-
Epistatic interactions: Detecting and characterizing epistatic interactions between QTLs stays a major problem.
-
Gene × surroundings interactions: Accounting for the complicated interactions between genes and the surroundings is essential for correct QTL mapping.
-
Low decision mapping: The decision of QTL mapping is commonly restricted, making it troublesome to pinpoint the precise location of QTGs.
-
A number of testing correction: The necessity to right for a number of testing when analyzing many markers can scale back the facility of QTL detection.
Future instructions in QTL mapping embody:
-
Improvement of extra highly effective statistical strategies: Improved strategies are wanted to deal with complicated knowledge units and account for epistatic and GxE interactions.
-
Integration of genomic info: Combining QTL mapping with genomic sequencing knowledge can enhance the decision of QTL mapping and establish candidate genes.
-
Software of high-throughput phenotyping applied sciences: Excessive-throughput phenotyping permits for the gathering of enormous quantities of phenotypic knowledge, rising the facility of QTL mapping research.
In conclusion, QTL mapping is a priceless software for dissecting the genetic structure of complicated traits. Whereas challenges stay, ongoing developments in statistical strategies, genomic applied sciences, and high-throughput phenotyping are regularly bettering the facility and backbone of QTL mapping, resulting in a deeper understanding of the genetic foundation of complicated traits throughout various organic programs.





Closure
Thus, we hope this text has offered priceless insights into Mapping Quantitative Trait Loci (QTL): Unraveling the Genetic Structure of Complicated Traits. We hope you discover this text informative and helpful. See you in our subsequent article!